Do you find it hard at times to get into a daily rhythm or routine? You may have heard of our circadian rhythm also coined as an organism’s “internal clock,” in which the body’s routines are regulated on a system that runs roughly 24hrs (oh wouldn’t you know that’s the length of a day!). It has been known to be monitored by the environment such as light and temperature. This doesn’t only have to do with sleep but also includes the molecular processes during sleep and wakefulness that keep an organism in homeostasis. It is safe to say that imbalances with this system can greatly impact an organism. For example, if we think of the opening and closing of the vascular plants stomata which is dependent on light (ie. daytime/nighttime) and temperature and that it is allowing for optimal energy consumption and storage this is a prime example that plants 24-hour clock needs to remain on its homeostatic schedule. Additionally several genetic diseases have been linked to malfunctions in alternative splicing (ex. tumor progression, ie. cancers), which supports how detrimental the impact improper circadian rhythm can have on health.
As it turns out, there is more than just one genetic clock that organisms harness. There is the brain’s Suprachiasmatic nucleus (SCN), known as the ‘Master Clock’, the pacemaker for an organism on a full-body scale, mainly regulating wake and sleep. What may be lesser known is that there are also the peripheral circadian rhythms, being tissue-dependent, are regulating the same genes as in other organ tissues in different ways. These messages may not always go up to the big clock in the brain but are still regulating gene expression post-transcriptionally.
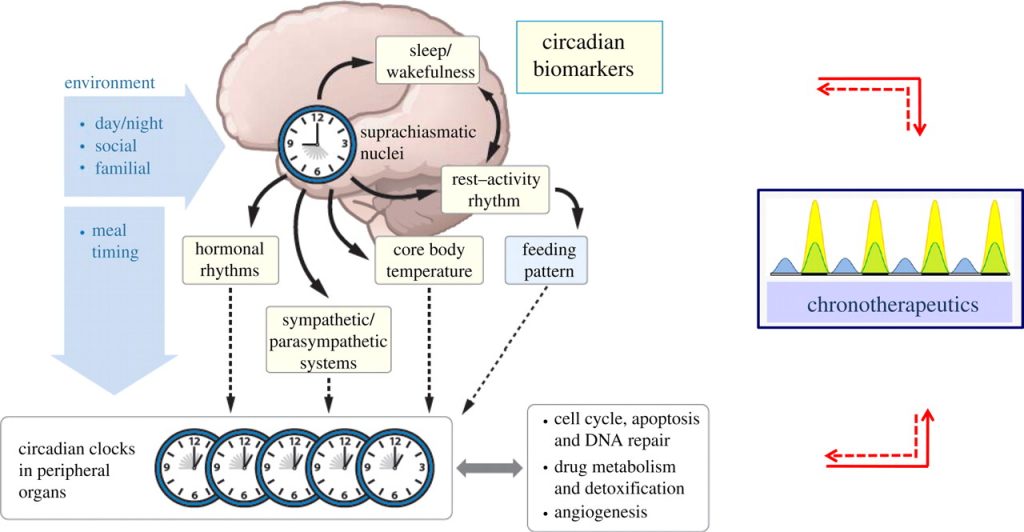
Outline of the circadian rhythm timing controlled largely by the Suprachiasmatic Nucleus (SCN) in the brain. Image retrieved from: http://rsfs.royalsocietypublishing.org/content/1/1/48
The curiosity that comes with this transcription/translation feedback has led to the discovery of a biochemical and genetic basis for the mechanism that is the 24-hour clock. The main component being and RNA negative feedback loop of proteins in which the protein itself when produced hinders its own expression. There are genes known as FREQUENCY (fre), PERIOD (per), and TIMLESS (tim) involved in circadian control in which oscillation in their expression impacts the clock. This oscillation in mRNAs is what has lead researchers towards how the mechanics of splicing are associated with circadian rhythm and its functioning. Splicing (occurring only in eukaryotes) is essentially the cleavage and assortment of intron/exon containing pre-mRNA to form exon-only mature mRNA so that translation may occur. This process can produce a myriad of genetic coding from only one piece of pre-mRNA in which splicing can be a selective process under certain given conditions including those environmental factors which help regulate circadian rhythm (ex. night and day).
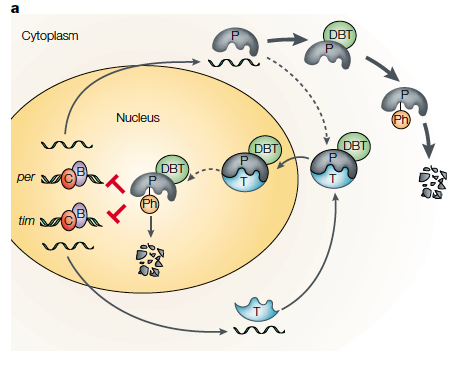
Top: example of per and tim gene cycle (in Drosophila melanogaster (fruit fly) where their production is regulated by a negative feedback loop. Bottom: the fluctuation of circadian genetic transcripts in a 24 hour day (white bar represents light, black is darkness). PER and CLK are proteins which regulate circadian rhythm processes (such as metabolism) whose expression varies throughout the day. For more background the figures obtained from a peer reviewed journal giving an outline of the circadian rhythm of multiple organisms: http://www.nature.com/nrg/journal/v2/n9/abs/nrg0901_702a.html
A lot of research has continuously supported the fact that circadian rhythm is controlled on a molecular level (with transcription and translation factors), what has more recently come to light is the effects circadian rhythm has on molecular controls and post-transcriptional mediation. This mediation has been demonstrated to be tissue-dependent and time sensitive. Studies have been presenting much evidence towards what mechanisms relate post-transcriptional regulation and the circadian rhythm, which includes alternative splicing. Although it has been speculated that some transcriptional-translational processes are not driven by the circadian rhythm there are those which are greatly impacted on a peripheral scale. These changes in an organism’s environment promote timely oscillations in individual cells depending on what is required at a certain time (ie. does the organism need to digest food, sleep or be active?).
In 2012 McGlincy et. al investigated the potential relationship between the circadian rhythm and alternative splicing in that alternative splicing of certain genes are regulated by it. Alternative splicing increases a single genes capacity to impact bodily function. For example, Dscam which contains 95 alternate cassette exons has the potential to encode 38,016 different isoforms! The exons which are included in any mature mRNA are very specific to the protein they will translate and so any variation has a large impact. As such, McGlincy et. al found that although every cell has the same amount of genetic information, the same gene may splice differently and be spliced according to the specific tissues needs. This study recognized that there are clear differences in expression between day and night by measuring the rate of alternative splicing and transcription expression to reveal that some genes displayed a temporal relationship between the two. It was also found that single genes were integrating circadian signals at different levels and that fasting causes changes in alternative splicing patterns. This change in alternative splicing patterns thus leads to incorporation of different processes and these processes happening at different times and frequencies which when a mutation occurs can
First bit more background on the circadian rhythm transcription factors, there is CLOCK and BMAL1 which are at the basis of circadian rhythm gene expression. The negative feedback loop of clock genes such as period (per) and cryptochrome (cry) genes in which their translocation back into the nucleus represses their activation (ie. negative feedback loop). This intrigue of how alternative splicing can occur due to environmental and thus circadian change has promoted others to investigate. Preubner et. al in 2014 studied how this relationship of post-transcriptional regulation and circadian rhythm is more than just feedback loops and can be explained by observing when alternative splicing occurs. They were interested in splicing factor U2af26 which encodes a splicing protein. Alternative splicing of this protein, under given circadian conditions, causes a frameshift where 143 new amino acids are coded for. This alternatively spliced mRNA showed to destabilize PER1 protein (a protein coded by period gene) hindering the negative feedback loop required for homeostatic rhythm.
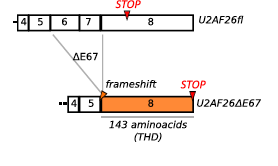
Schema of the frameshift that occurs when U2af26 is alternatively spliced. This causes exon 6 and 7 to not be ligated into the mature mRNA and exon 8 becomes fully available. THD stands for Timeless homology domain which is a homologue to Drosophila melanogaster’s timeless gene which is why this alternatively spliced gene is believed to be related to circadian rhythm control. Retrieved from: http://www.cell.com/molecular-cell/abstract/S1097-2765(14)00326-8?_returnURL=http%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS1097276514003268%3Fshowall%3D
This effect that the alternatively splice U2af26 gene is not completely random as the new 143 amino acid sequence now being coded is a homologue to the fruit flies Timeless gene which is yet another regulator of circadian rhythm. Further promoting the connection between circadian rhythm and molecular processes more specifically splicing.

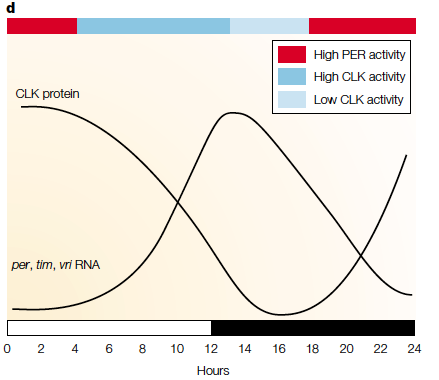
Comparison between the alternatively spliced U2AF26 gene (renamed U2AF2667 as exons 6 and 7 are removed) in mice and the TIM gene in Drosophila, both having an impact on circadian rhythm oscillations. Retrieved from Preubner et. al (2014): http://www.cell.com/molecular-cell/abstract/S1097-2765(14)00326-8?_returnURL=http%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS1097276514003268%3Fshowall%3D
It has become clear to many that it is important to continue to dig into the unknown mechanisms of the circadian rhythm as it greatly impacts an organism’s adaptation to its environment and possible changes in its metabolic conditions. The research that has been conduction is only the beginning of a large stream of investigations to get to the basis of this intricate relationship. It is indeed important to continue with these studies so that many diseases involving an improper alignment of cell processes can receive more advanced treatment. Talk about an integrated system!


Recent Comments